Atsushi Matsuzawa
Development of therapeutic strategies for various diseases by regulation of the cell death/survival balance using ubiquitin chemo-technologies
 |
Atsushi Matsuzawa, PhDLaboratory of Health Chemistry, Graduate School of Pharmaceutical Sciences, Tohoku University |
|---|
Research summary
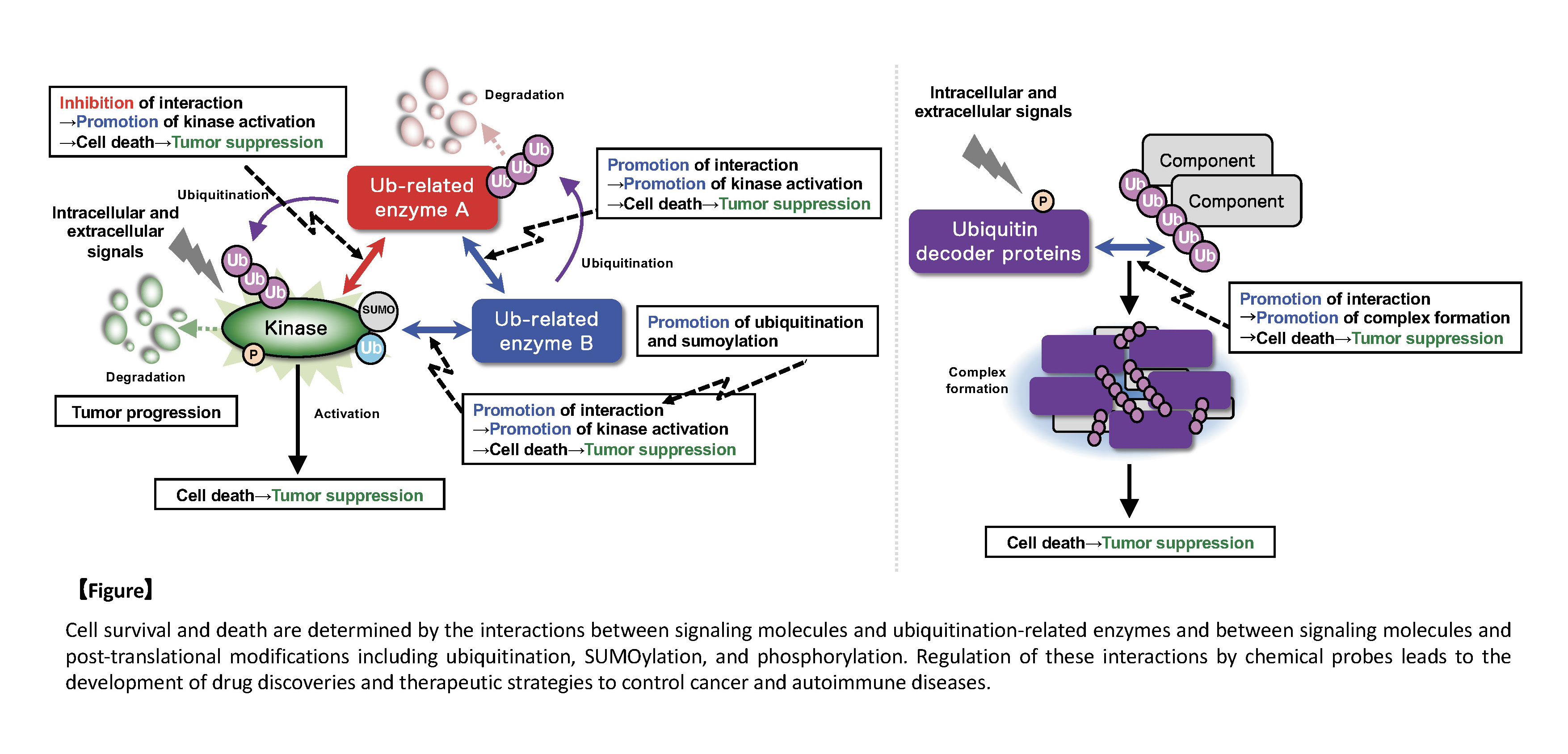
Cellular responses, such as cell viability, are determined by the balance of various signals, which are inputted from the inside and outside of the cells and regulate cell proliferation, differentiation, and death. Recently, we found that multiple ubiquitination-related enzymes clustered around signaling molecules regulating cell life and death, formed the complex, and controlled their activities each other, and that the specific complex consisting of ubiquitin and ubiquitin decoder proteins is also formed, in order to fine-tune the signal balance, which determines cell viability and induces appropriate cellular responses. When the signal balance is disrupted, abnormal signals are generated and induce excessive cell death and proliferation, leading to cancer and autoimmune diseases. We also found that post-translational modifications, such as ubiquitination, SUMOylation, and phosphorylation, are essential for the interaction between multiple components in the complex, and therefore, the regulation of these modifications suppresses excessive cell death and proliferation and enables the treatment of various diseases, including cancer and autoimmune diseases, which are induced by the imbalance in the signals regulating cell life and death.
For example, a kinase that regulates cell viability forms a complex together with different ubiquitination-related enzymes, which control the activity of the kinase. One ubiquitination-related enzyme is directly involved in ubiquitination-dependent degradation of the kinase, and another ubiquitination-related enzyme contributes to ubiquitination-dependent degradation of the former ubiquitination-related enzyme, leading to indirect regulation of the kinase. Thus, such the complicated fine-tuning mechanism exists in the signals. In addition, the formation of the complex consisting of ubiquitin and ubiquitin decoder proteins in response to stress stimuli is critical for the induction of atypical cell death. The interaction between these molecules is found to require post-translational modifications, such as ubiquitination. In other words, it is important to elucidate the mechanisms of the complex formation and the regulation of the signal balance determining cell life and death at the molecular level, because the interaction between signaling molecules and the recognition of post-translational modifications by decoder proteins are essential points (sites of action) for the regulation of the signal balance determining cell life and death. Therefore, we believe that these points can be artificially controled and fine-tuned by chemotechnology at the molecular level.
In this study, we would like to reveal that the controled signal balance of cellular responses, such as cell life and death, leads to the suppression of excessive cell death and proliferation, through appropriate regulation of the positive or negative interaction between ubiquitination-related modifications and signaling molecules, using chemotechnology, and to propose novel therapeutic strategies for cancer and immune diseases, which are different from conventional methods.
Publications
- Toyama T, Hoshi T, Noguchi T, Saito Y, Matsuzawa A, Naganuma A, *Hwang G W.
Methylmercury induces neuronal cell death by inducing TNF-α expression through the ASK1/p38 signaling pathway in microglia.
Sci. Rep. 11, 9832 (2021)
PMID: 33972601 - Hirata Y, Takahashi M, Yamada Y, Matsui R, Inoue A, Ashida R, Noguchi T, *Matsuzawa A.
trans-Fatty acids promote p53-dependent apoptosis triggered by cisplatin-induced DNA interstrand crosslinks via the Nox-RIP1-ASK1-MAPK pathway.
Sci. Rep. 11, 10350 (2021)
PMID: 33990641 - Shimada T, Kudoh Y, Noguchi T, Kagi T, Suzuki M, Tsuchida M, Komatsu H, Takahashi M, Hirata Y, *Matsuzawa A.
The E3 Ubiquitin-Protein Ligase RNF4 Promotes TNF-α-Induced Cell Death Triggered by RIPK1.
Int. J. Mol. Sci. 22, 5796 (2021)
PMID: 34071450 - Kagi T, Naganuma R, Inoue A, Noguchi T, Hamano S, Sekiguchi Y, Hwang G W, Hirata Y, *Matsuzawa A.
The polypeptide antibiotic polymyxin B acts as a pro-inflammatory irritant by preferentially targeting macrophages.
J. Antibiot. 75, 29-39 (2022)
PMID: 34824374 - Shimada T, Yabuki Y, Noguchi T, Tsuchida M, Komatsu R, Hamano S, Yamada M, Ezaki Y, Hirata Y, *Matsuzawa A.
The distinct roles of LKB1 and AMPK in p53-dependent apoptosis induced by cisplatin.
Int. J. Mol. Sci. 23, 10064 (2022)
PMID: 36077459 - Lee J Y, Kim J M, Noguchi T, Matsuzawa A, Naganuma A, *Hwang G W.
Deubiquitinase USP54 attenuates methylmercury toxicity in human embryonic kidney 293 cells.
Fudam. Toxicol. Sci. 9, 159-162 (2022) - Luo Y R, *Kudo T A, Tominami K, Izumi S, Tanaka T, Hayashi Y, Noguchi T, Matsuzawa A, Nakai J, Hong G, Wang H.
SP600125 enhances temperature-controlled repeated thermal stimulation-ynduced neurite outgrowth in PC12-P1F1 cells.
Int. J. Mol. Sci. 23, 15602 (2022)
PMID: 36555248 - Sekiguchi Y, Takano S, Noguchi T, Kagi T, Komatsu R, Tan M, Hirata Y, *Matsuzawa A.
The NLRP3 inflammasome works as a sensor for detecting hypoactivity of the mitochondrial Src family kinases.
J. Immunol. (2023)
PMID: 36744909
Former Publications
- Matsuzawa A, Tseng PH, Vallabhapurapu S, Luo JL, Zhang W, Wang H, Vignali DA, Gallagher E, *Karin M.
Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex.
Science (Research Article) 321, 663-668 (2008)
PMID: 18635759 - Hirata Y, Katagiri K, Nagaoka K, Morishita T, Kudoh Y, Hatta T, Naguro I, Kano K, Udagawa T, Natsume T, Aoki J, Inada T, Noguchi T, Ichijo H, *Matsuzawa A.
TRIM48 promotes ASK1 activation and cell death through ubiquitination-dependent degradation of the ASK1 negative regulator PRMT1.
Cell Rep. 21, 2447-2457 (2017)
PMID: 29186683 - Noguchi T, Suzuki M, Mutoh N, Hirata Y, Tsuchida M, Miyagawa S, Hwang G W, Aoki J, *Matsuzawa A.
Nuclear-accumulated SQSTM1/p62-based ALIS act as microdomains sensing cellular stresses and triggering oxidative stress-induced parthanatos.
Cell Death Dis. 9, 1193 (2018)
PMID: 30546061 - Hirata Y, Inoue A, Suzuki S, Takahashi M, Matsui R, Kono N, Noguchi T, *Matsuzawa A.
trans-Fatty acids facilitate DNA damage-induced apoptosis through the mitochondrial JNK-Sab-ROS positive feedback loop.
Sci. Rep. 10, 2743 (2020)
PMID: 32066809 - Noguchi T, Sekiguchi Y, Kudoh Y, Naganuma R, Kagi T, Nishidate A, Maeda K, Ishi C, Toyama T, Hirata Y, Hwang GW, *Matsuzawa A.
Gefitinib initiates sterile inflammation by promoting IL-1β and HMGB1 release via two distinct mechanisms.
Cell Death Dis. 12, 49 (2021)
PMID: 33414419