Yusuke Sato
Structural biology of branched ubiquitin chain recognition using enzymatic and chemical reactions
 |
Yusuke Sato, PhDTottori University, Graduate School of Engineering, Laboratory of Structural Biology |
|---|
Research summary
Protein ubiquitination is a post-translational modification that regulates various biological processes. Ubiquitin is typically conjugated to the amino group of the substrate lysine residue via its C-terminal carboxyl group. The amino groups of the N-terminal methionine residue and seven lysine residues of ubiquitin can be covalently bonded to the C-terminus of another ubiquitin to form eight types of polyubiquitin chains (M1, K6, K11, K27, K29, K33, K48, K68 and M1). Ubiquitin chains have different conformations depending on the residues used for the linkage and are recognized by different decoder molecules, so ubiquitin modification can regulate a wide variety of biological processes. Structural studies have demonstrated the recognition mechanisms of specific ubiquitin chains by decoder and eraser molecules. On the other hand, the specific recognition mechanism of branched chains by decoder and eraser molecules has not yet been elucidated, although the function of branching chains in the cell has been gradually reported and their importance has been pointed out.
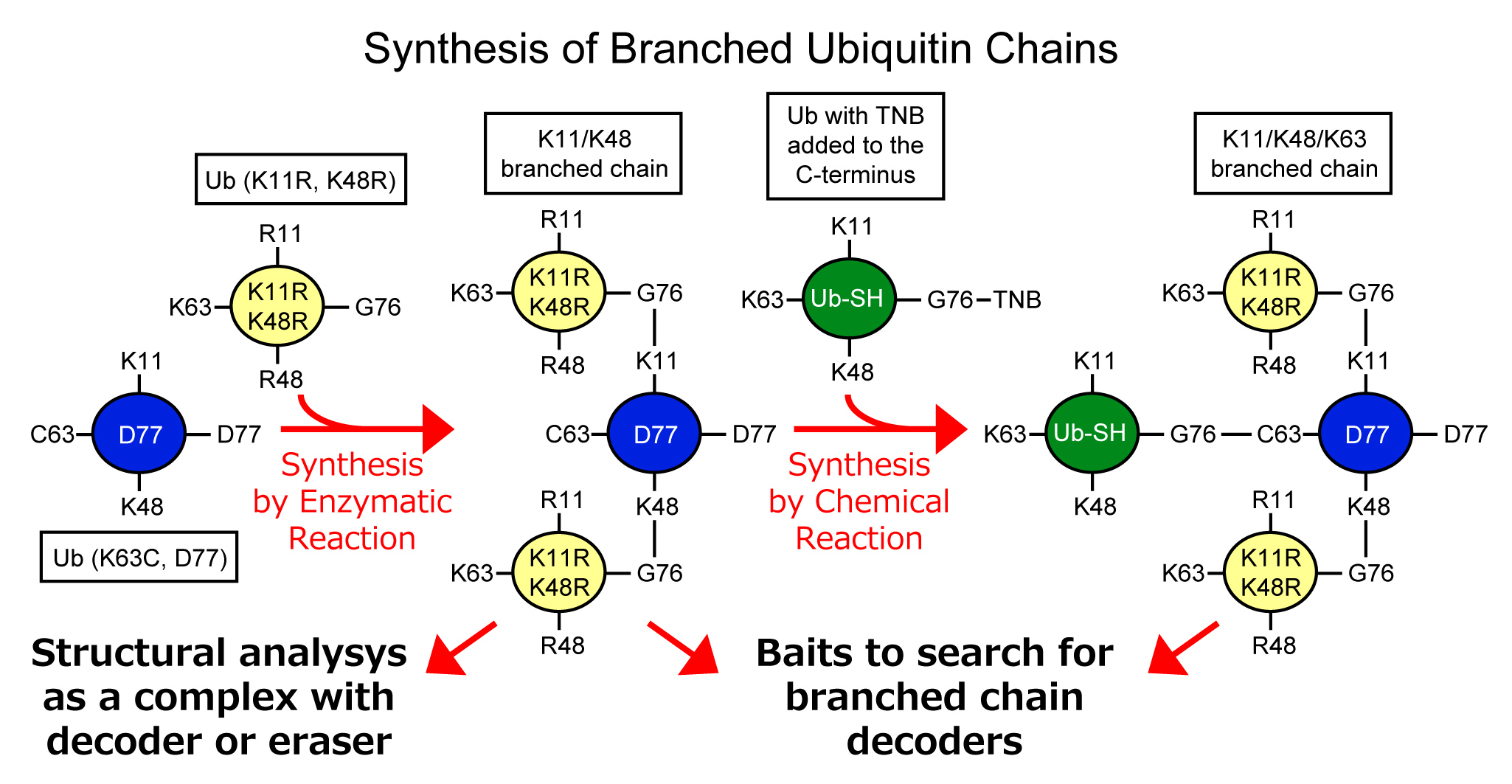
In this study, we will establish a method to synthesize a large amount of branched chains by combining enzymatic and chemical reactions, and determine the crystal structure of branched chains in complex with decoder or eraser molecules. It is possible to synthesize branched chains using combinations of E2 and E3 enzymes that has linkage specific elongation activity. However, it is difficult to control the position of the branch and the length of the ubiquitin chain when using wild-type ubiquitin. Therefore, we will use ubiquitin with lysine mutants to prevent unwanted ubiquitin chain extension. On the other hand, in some branching combinations, the previously bound ubiquitin inhibits the approaching of enzymes and make it difficult for enzymatic elongation of branched chains. In such cases, we plan to use a chemical reaction method based on disulfide bonds to synthesis branched chains which is difficult to make by enzymatic reaction. Synthesized branched chains will be possible to use for structural analysis, and as baits for searching for decoder molecules of branched chains.
Publications
- Li Y, Okatsu K, Fukai S, *Sato Y.
Structural basis for specific recognition of K6-linked polyubiquitin chains by the TAB2 NZF domain.
Biophys J. 120, 3355-3362 (2021)
PMID: 34242591 - *Nakazawa S, Mamiya R, Kawabata-Iwakawa R, Oikawa D, Kaira K, Tokunaga F, Nobusawa S, Sato Y, Sasaki A, Yajima T, Shirabe K.
Identification and molecular analysis of RNF31 Q622H germline polymorphism.
Oncol Lett. 24, 394 (2022)
PMID: 36276481 - Saito R, Tada Y, Oikawa D, Sato Y, Seto M, Satoh A, Kume K, Ueki N, Nakashima M, Hayashi S, Toyoshima Y, Tokunaga F, Kawakami H, *Kakita A.
Spinocerebellar ataxia type 17-digenic TBP/STUB1 disease: neuropathologic features of an autopsied patient.
Acta Neuropathol Commun. 10, 177 (2022)
PMID: 36476347
Former Publications
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, *Fukai S.
Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains.
Nature 455, 358-362 (2008)
PMID: 18758443 - Sato Y, Fujita H, Yoshikawa A, Yamashita M, Yamagata A, Kaiser SE, *Iwai K, *Fukai S.
Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex.
Proc. Natl. Acad. Sci. U.S.A. 108, 20520-20525 (2011)
PMID: 22139374 - Sato Y, Goto E, Shibata Y, Kubota Y, Yamagata A, Goto-Ito S, Kubota K, Inoue J, Takekawa M, Tokunaga F, *Fukai S.
Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity.
Nat. Struct. Mol. Biol. 22, 222-229 (2015)
PMID: 25686088 - Sato Y, Okatsu K, Saeki Y, Yamano K, Matsuda N, Kaiho A, Yamagata A, Goto-Ito S, Ishikawa M, Hashimoto Y, Tanaka K, *Fukai S.
Structural basis for specific cleavage of Lys6-linked polyubiquitin chains by USP30.
Nat. Struct. Mol. Biol. 24, 911-919 (2017)
PMID: 28945247 - Sato Y, Tsuchiya H, Yamagata A, Okatsu K, Tanaka K, *Saeki Y, *Fukai S.
Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat. Commun. 10, 5708 (2019)
PMID: 31836717