Satoshi Yamanaka
Development of PROTAC-dependent interactome analysis method using proximity-dependent biotinylation enzyme
 |
Satoshi Yamanaka, PhDEhime University, Proteo-Science Center, Division of Proteo-Interactome Assistant professor |
|---|
Research summary
Small molecule compounds that induce the degradation of target proteins, such as thalidomide and its derivatives (Immunomodulatory Drugs/IMiDs), are called “protein degrader”. Protein degraders are attractive drugs for the treatment of many diseases and functional analysis of target proteins because of their powerful action which induces the degradation of target proteins thoroughly. Furthermore, today, they is applied to PROTAC (Proteolysis targeting chimera) or SNIPER (Specific and Nongenetic IAP-dependent Protein Eraser dependent), which is heterobifunctional protein degrader consisting of a ligand on an E3 ubiquitin ligase (E3 binder) such as IMiDs, a linker, and a ligand on target proteins (Target binder). Analysis of PROTAC- or SNIPER-dependent interacting proteins with E3 ubiquitin ligase is an important research field to understand the actions of PROTAC or SNIPER.
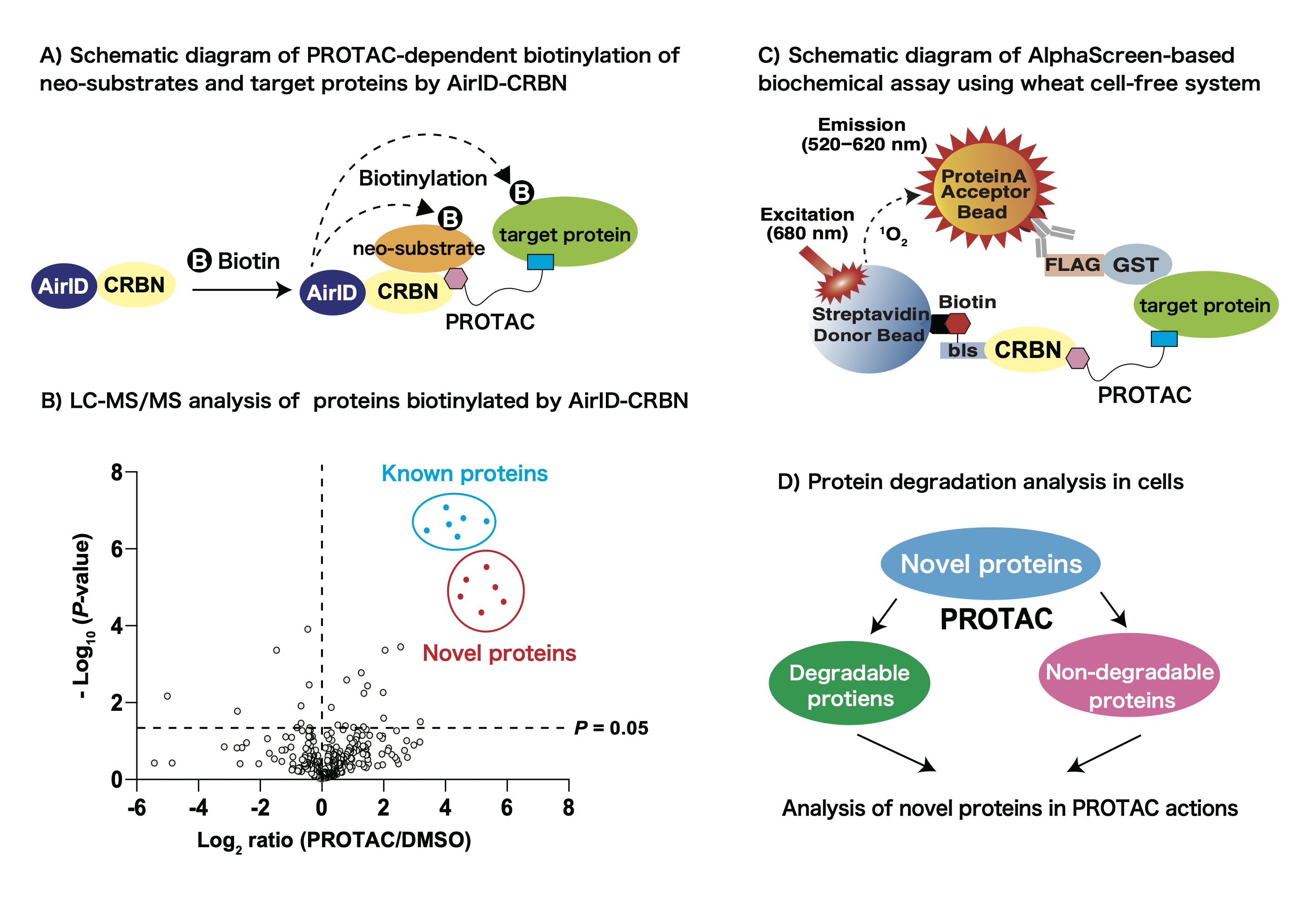
So far, we have developed a novel proximity-dependent biotin identification enzyme (Ancestral BirA for proximity-dependent biotin identification/AirID) as a useful enzyme for protein-protein interactions. In addition, we reported that LC-MS/MS analysis of biotinylated peptides using AirID enabled us to detect protein-protein interaction. In this study, we aim to develop a PROTAC-dependent interactome analysis method using AirID-fused CRBN, which is a substrate recognition receptor in cullin E3 ligase complex (Fig. A). Firstly, we plan to investigate the optimal conditions for biotinylation of PROTAC-dependent target proteins by AirID-CRBN. Then, we plan to perform LC-MS/MS analysis of biotinylated peptides in a PROTAC-dependent manner (Fig. B). Next, we plan to examine whether the identified proteins interact with CRBN in a PROTAC-dependent manner (Fig. C) and whether the identified proteins are degraded in cells (Fig. D). By performing these analyses, we aim to develop PROTAC-dependent interactome analysis method to understand the actions of PROTAC.
Publications
- Yamanaka S, Horiuchi Y, Matsuoka S, Kido K, Nishino K, Maeno M, Shibata N, Kosako H, *Sawasaki T.
A proximity biotinylation-based approach to identify protein-E3 ligase interactions induced by PROTACs and molecular glues.
Nature Communications 13, 183 (2022)
PMID: 35013300
Former Publications
- Yamanaka S, Sato Y, Oikawa D, Goto E, Fukai S, Tokunaga F, *Takahashi H, *Sawasaki, T. Subquinocin, a small molecule inhibitor of CYLD and USP-family deubiquitinating enzymes, promotes NF-κB signaling.
- Kido K, Yamanaka S, Nakano S, Motani K, Shinohara S, Nozawa A, Kosako H, Ito S, *Sawasaki T. AirID, a novel proximity biotinylation enzyme, for analysis of protein-protein interactions.
- Yamanaka S, Shoya Y, Matsuoka S, Nishida-Fukuda H, Shibata N, *Sawasaki T. An IMiD-induced SALL4 degron system for selective degradation of target proteins.
- Furihata H, Yamanaka S, Honda T, Miyauchi Y, Asano A, Shibata N, *Tanokura M, *Sawasaki T, *Miyakawa T. Structural bases of IMiD selectivity that emerges by 5-hydroxythalidomide.
- Yamanaka S, Murai H, Saito D, Abe G, Tokunaga E, Iwasaki T, Takahashi H, Takeda H, Suzuki T, Shibata N, Tamura K, *Sawasaki T. Thalidomide and its metabolite 5-hydroxythalidomide induce teratogenicity via the cereblon neosubstrate PLZF.
Biochem. Biophys. Res. Commun. 524, 1-7 (2020)
PMID: 31898971
eLife 9, e54983 (2020)
PMID: 32391793
Commun. Biol. 3, 515 (2020)
PMID: 32948804
Nat. Commun. 11, 4578 (2020)
PMID: 32929090
EMBO J. 40, e105375 (2021)
PMID: 33470442